Archive : Article / Volume 2, Issue 1
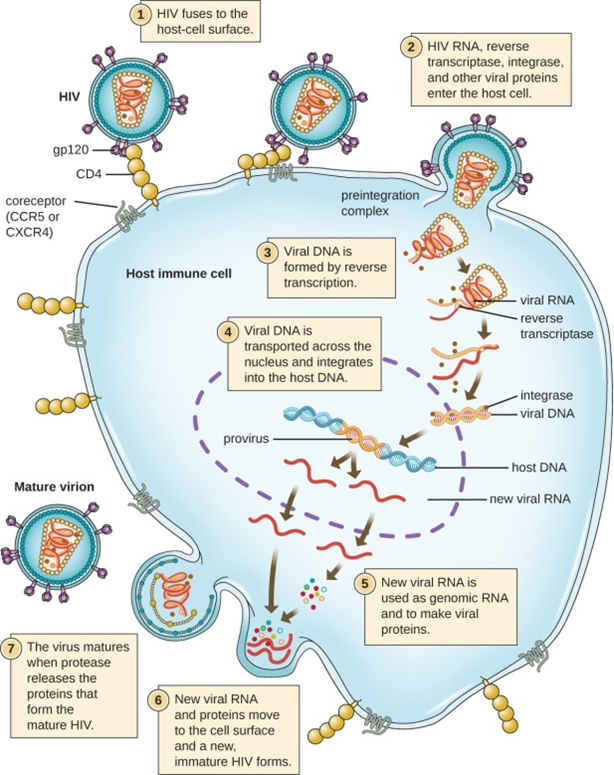
Case Report | DOI: https://doi.org/10.58489/2836-5003/008
Transmitting, mutational and pandemic nature of corona: the role of transposons, is1: tn:is1 and crispr- cas-9
Ex Faculty IIT-Kharagpur.721302, India, IGE (Institute of Genetic Engineering) Badu- Kolkata, India
Correspondng Author: Nitosh Kumar Brahma
Citation: Nitosh Kumar Brahma (2023) Transmitting, mutational and pandemic nature of corona: the role of transposons, is1: tn:is1 and crispr- cas-9. Archives of Immunology Research and Therapy.2(1). DOI: 10.58489/2836-5003/008
Copyright: © 2023 Nitosh Kumar Brahma, this is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received Date: 2023-01-18, Received Date: 2023-01-18, Published Date: 2023-01-30
Abstract Keywords: transposons, alveoli, transcription, translation, mutation and infections.
Abstract
Corona/ Covid19/Omicron / SARS/ MARS similar viruses have no individual genetic mechanism to make them encapsulated spore forming microbes similar to bacteria, fungi, algal blooms involved spreading allergy. Transmission in all such cases need fluid dynamics of water and air. This flow does not require any support of matrix, i.e., nano particles of carbon, silicates and dust of animate inanimate molecules. They enter into lung alveoli surface epithelial cell, adhere on the surface of body saliva, originated due to cold and other infections. In support of ACE-I and II (Angiotensin Converting Enzyme I and II). So, there are two releasing mechanisms are existing in corona infection; one is the release of corona virus from the alveoli cells with matrix binds of one co-morbid patients directly to other healthy recipient at the distance of 6 m in a closed environment by its self-development irritation and allergy, as received by sudden irritating smoke developed from kitchen, pollen, that grows during grass cutting, cleaning and sanitization. Transcription of m-RNA, in a matrix based single corona after entering into lung alveoli starts to transcribe and translate to many molecules of corona viruses, damages alveoli cells of lung, caused due to individual physiological conditions, old (above 70 years) and young (below 40), co-morbid(sick) and healthy people at similar age groups. IS: Tn:IS), Crispr/Cas-9 DNA transposons existing in corona m-RNA starts to act during transcription, and translation
Introduction
An AAIR( Ant adherent Immune Response) experimental was initiated in Bulb/C mic against serotype 026: EPEC (Enteropathogenic (invasive) Escherichia coli) a fatal diarrheagenic E.coli , killing mice over night at ( 8 hours) found unique support of hybrid genetically engineered E.coli, carrying MRHU(+) donor plasmid in support of trascojugated pRT ( Plasmid Ronald Taylor)of Yale University , USA, 1980, expressed MRHU(+) plasmid in auxotrophic genetically Engineered (GE) Immune responses, revealed that the mice were resistant against the donors 026:EPEC, even at highest dose of 10^8 cells / 0.2 ml, intraperitoneal inoculum, without pre-inoculated Balb/ C mice were killed at low intra peritoneal dose of 101 cells / 0.2 ml. The molecular investigation revealed that some IS based transposons support the expression of MRHU (+) hemagglutination in hybrid genes of Escherichia coli. By increasing antibiotic profiles, as occurred due to IS (Insertion Sequence) as represented by IS1, IS2, IS3, -.IS10 DNA sequence change the flanking nature of transposons (Tn) “IS: Tn:IS” through antibiotic resistance profile initiate illegitimate recombination and was carefully studied in 1980 in the lab of Prof. H. Saedler, Biology-III Genetic Engineering, ALU, Freiburg, Germany. In case of increasing fatality of corona antibiotic treatments is recommended.
Considering the fatality rate of corona as pandemic globally, the author has attempted to recapitulate the possible application of AAIR, IS: Tn:IS and Crispr/Cas-9 in designing vaccine against corona, to prevent corona virus to adhere on trachea and lung cells, to repair and to bring healthy life to mankind. IS: Tn:IS was observed/studied/discovered in Maize by Barbara Mc Clintock, USA, Peter Starlinger and Heinz Saedler, Germany studied the same in bacteria. Due to spontaneous mutations in corona, it is speculated by the author, that “IS: Tn:IS” DNA sequences might be present in corona, to mutate and to change the adhering, invasive and infective spike protein. The presence of IS-Tn-IS sequence have not been studied in SARS-COV-2, COVID-19, and corona. The sequence length, varied from IS1 (0.8kb) to IS10, (1.2 kb) (kilo base pairs) showed the potentiality spontaneous to do illegitimate recombination. If any of the sequence IS1, IS2, IS3…IS10 found common among HIV, MERS, SARS, Influenza, and H1N1 viruses, it could be useful to develop AAIR. Both AAIR vaccine concept and recent revolution in DNA editing, Crispr/cas-9(Clustered, Repeat Interspaced Short Palindromic Repeat), / (Crispr-associated protein-9) both could essentially be used, bidirectional in corona vaccine to protect pandemics and their repeats in future.
The IR mechanisms which are proposed is supposed to be controlled by IS: Tn:IS transposons failed to attach the spike proteins. Hybrid DNA as involved in formation of AAIR as monoclonal antibody will block the adherence of corona spikes. Crispr/cas-9 will repair in addition to the transmitting, invasive nature of corona. In the process the programmable DNA of non-pathogenic influenza virus. The author describes few of those possible mechanisms of Crispr transposons involved in changing protein structures of spike protein of corona. Jumping gene, transposons, if programmed non-pathogen m-RNA could be used in the form of vaccine. Bacteriophage Lambda, x similar and other fusion to change the mode of infection to coccucea are the present thought, changing the direction of corona to human trillion microbes involved in Pneumonia
Materials and Methods
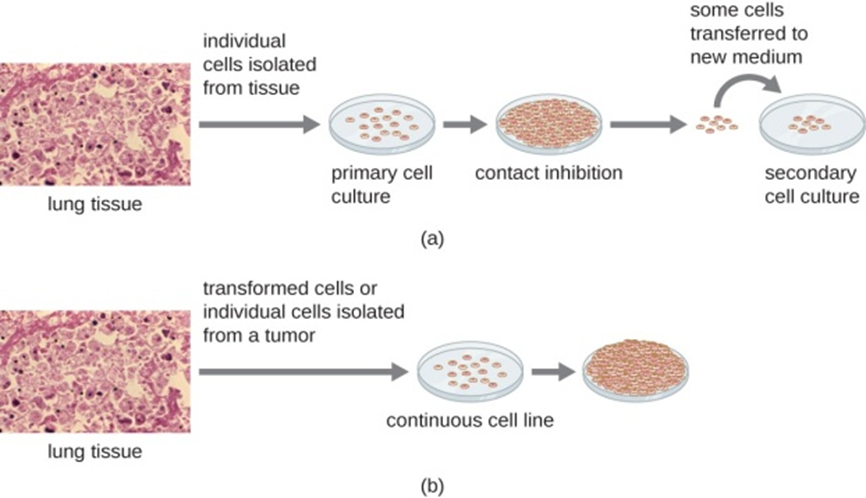
The study inspired not only to study genetic rearrangements and illegitimate recombination, as occurred when corona infections are moving downwards (i.e., many varieties of antibiotics are applied, various sanitations are used in commonplaces, when for survivals corona stats to mutate and exposed as antibiotic resistant human system surviving with bacterial strains, namely pneumococcal strains significantly participate the micro evolution of living world. Fig.(d), represents the ampicillin resistant Tn. Fig.1 (e), shows the segregating colour changes among maize seeds, as caused by transposons. Fig.1 (f) is reproduced with the legendary personality of Barbara Mc Clintock, who identified colour changes in maize seeds due to presence of spm transposons in maize seeds DNA in 1931, and received Nobel Prize in 1983, [1].
IS based Tn (Transposons) are involved in higher and lower groups of plant and animal kingdom, including the evolution of viruses and bacteria. The camouflaging infective nature of SARS COV-2 corona virus, covid-19 could be associated with Tn, as proposed by the author, [10-14]. Jumping-genes as retro transposons [9] hijacks special cells called nurse cells, produce invasive nature of DNA driving evolution, and causing diseases.
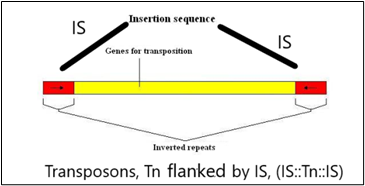
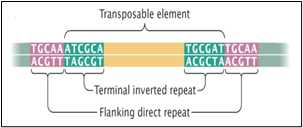
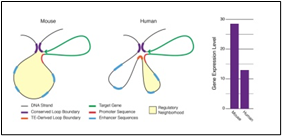


Fig.1 (a) shows one transposon, flanked by IS elements (IS: Tn:IS), Fig1(b) shows the repeats and inverted repeats DNA sequences involved of IS elements. Fig.1(C) shows the mechanism of Tn enzymes involved in illegitimate recombination and rearrangements. “Cut and Paste and Copy and Paste “foreign DNA have been identified in such illegitimate recombination where IS elements support to insert foreign DNA into host recipient DNA [1]. In Fig.1 (C). Histogram compares the chromatin folding frequencies among mouse and human. It shows that transposons-based chromatin folding in mouse is higher compared to human.IR (Immune Response) in mouse is stable, compared to human. MHC-HLA gene, responsible for IR in human chromosome 17 is susceptible compared to chromosome 6 in mouse [2-9] Chromatin loops are important for gene regulation because they define a gene's regulatory neighbor-hood, which contains the promoter and enhancer sequences responsible for determining its expression level. Remarkably, transposable elements (TEs) are responsible for creating around 1/3 of all loop boundaries in the human and mouse genomes, and contribute up to 75% of loops unique to either species. When a TE creates a human-specific or mouse-specific loop it can change a gene's regulatory neighborhood, leading to altered gene expression. The illustration shows a hypothetical region of the human and mouse genomes in which four enhancer sequences for the same target gene fall within a conserved loop. In this example, a TE-derived loop boundary in the human genome (orange bar) shrinks the regulatory neighborhood, preventing two of four enhancers from interacting with their target gene's promoter sequence. The net result is reduced gene expression in human relative to mouse. Looping variations such as these appear to be an important underlying cause of differential gene regulation across species and between different human cell types, suggesting that TE activity may play significant roles in evolution and disease. The method was studied by radio isotopic IS DNA labelling and southern hybridization. The method proves also how one IS elements jumps from plasmid to chromosome and back to plasmid. Almost half of our DNA sequences are made up of jumping genes, known as transposons. They jump around the genome in developing sperm and egg cells and are important in cellular evolution [15] and cause new mutations that lead to diseases. Remarkably little is known about when and where these movements started in development of reproductive cells. The key process that ensures their propagation for future generations with possible genetic disorders. Animals have developed a powerful using suppressing activities of jumping gene, non-coding pi RNAs [15-17], that recognize jumping genes and suppress their activity.
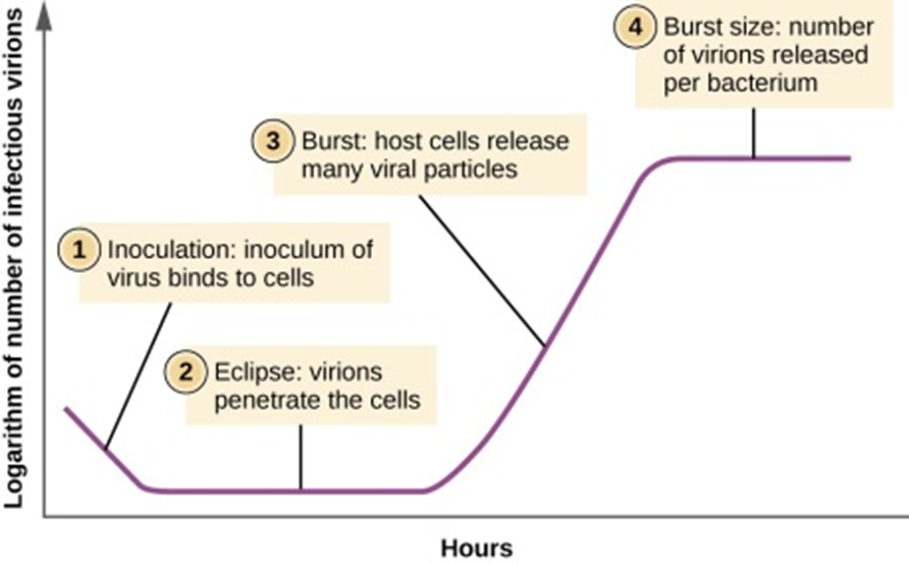
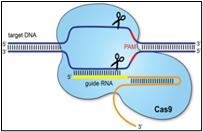
Some information has been recorded that virus, Covid-19, originated from Wuhan, China, and was spreaded all over the world, representing no RNA variations. Measurement of antibodies to SARS-CoV-2 will improve disease management, if used correctly. In late 2019, China reported a cluster of atypical pneumonia causative agent, responsible for severe acute respiratory syndrome (SARS), rapidly spreading across the globe through human interactions and respiratory transmissions and caused pandemic, Sequencing and searching the presence of IS elements in Covid-19 ss RNA are seems to be essential to understand pandemic and the delayed process of vaccination. 8 copies of IS1 and other number of copies of IS2, IS3 were identified in chromosomes and plasmid of Escherichia coli and other enteric infectious, water, soil and environment born. IS was also observed in plasmids of hybrid E. coli –K-12. It is obvious for the author that by observing the changing copies of IS significantly influenced the infective modes of bacteria, viruses and their evolution [10,14], compared to wild types that could have been widely used for the diagnosis of acute (current) SARS-CoV-2 infections [16,17]. To study and to compare the infective mode of corona and other SARS and MERS, H1N1, viruses, it is good that parallel to their phonotypical SEM (Scanning Electron Microscopy) views, molecular biology of those viruses are need to be studied by IS1 insertion sequence among all possible corona mutants, their wild types, among neumococcus and other related microbes, that predominate during infections and in increasing fatality. After an incubation period of 3 days, corona virus can cause the symptoms of a common cold, including nasal obstruction, sneezing, runny nose, secretions and coughing, [16-18].
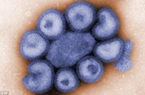
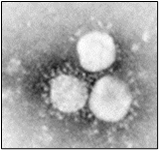
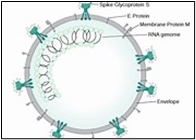
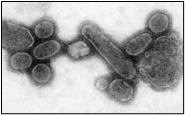
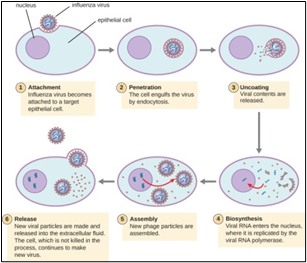
Fig.2, (a) show the SEM (Scanning Electron Microscope) views of H1N1, (b) Influenza virus colony, (c) SERS-COV-2 virus aggregating by their spike proteins in colony formation. The molecular structure of -COV-2 RNA-virus, ss RNA (Fig.2c1) transcribe coat- spike (S) glycoprotein, enzymatically initiate ACE-I, II (angiotensin converting enzyme, I ans II) surface activities of host cell, [19,20] to adhere membrane of lung cells. OC43 haemagglutinin type virions are released from host cells after rupturing respiratory cell membrane. New virions transmit the infection via airborne droplets to the healthy recipients. Virus replicates/multiply spontaneously in support of ss m-RNA and r-RNA of respiratory lung cells. (d) shows the outbreak of Spanish flu in 1918 pandemic. Viral clusters, and molecular biology significantly represent the similarities of their spike proteins, and colony formations. The envelope glycoprotein is the characteristic feature, the chemistry of adherence. By replicating and synthesising spike protein. Fig.2(e), shows the proposed cycle of m-RNA ss RNA Covid-19, that directly involved for rapid growths of spike (S) proteins, the capsid formation and multiplying virus particles in lung cells. 3’ ss m-RNA of Covid-19, recognizes 5’r-RNA of infected cells, translate glycoprotein, envelope membrane matrix protein and new corona virus particles, to spread infections.
Coronaviruses as reported found in avian and mammalian species, resemble morphologically-phenotypically similar, but differ by their antigenic structures. Even coronaviruses of human and cattle are found antigenically similar, varied by their pathogenic expression. As common colds symptom, respiratory infections, corona viruses, invade many different tissues and to initiate MOF (multiorgan failure) and fatal deaths of corona patients.
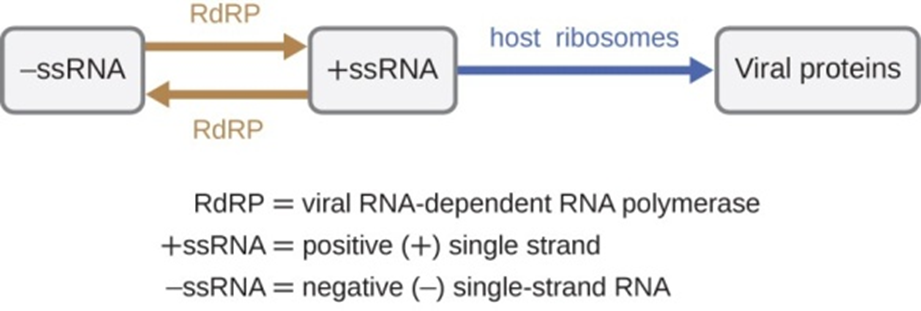
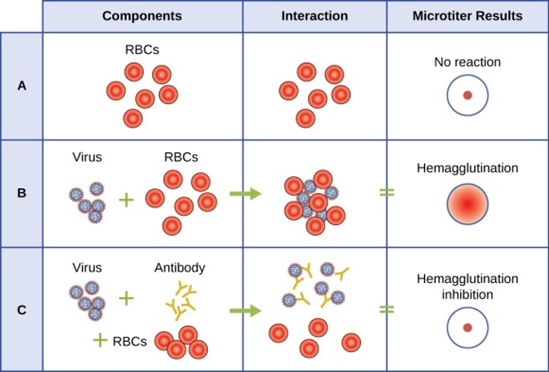
There are three types of RNA genome: dsRNA, positive (+) single-strand (+ssRNA) or negative (−) single-strand RNA (−ssRNA). If a virus has a +ssRNA genome, it can be translated directly to make viral proteins. Viral genomic +ssRNA acts like cellular mRNA. However, if a virus contains a −ssRNA genome, the host ribosomes cannot translate it until the −ssRNA is replicated into +ssRNA by viral RNA-dependent RNA polymerase (RdRP) (see Figure 5). The RdRP is brought in by the virus and can be used to make +ssRNA from the original −ssRNA genome. The RdRP is also an important enzyme for the replication of dsRNA viruses, because it uses the negative strand of the double-stranded genome as a template to create +ssRNA. The newly synthesized +ssRNA copies can then be translated by cellular ribosomes.
An alternative mechanism for viral nucleic acid synthesis is observed in the retroviruses, which are +ssRNA viruses (see Figure 6). Single-stranded RNA viruses such as HIV carry a special enzyme called reverse transcriptase within the capsid that synthesizes a complementary ssDNA (cDNA) copy using the +ssRNA genome as a template. The ssDNA is then made into dsDNA, which can integrate into the host chromosome and become a permanent part of the host. The integrated viral genome is called a provirus. The virus now can remain in the host for a long time to establish a chronic infection. The provirus stage is similar to the prophage stage in a bacterial infection during the lysogenic cycle. However, unlike prophage, the provirus does not undergo excision after splicing into the genome.
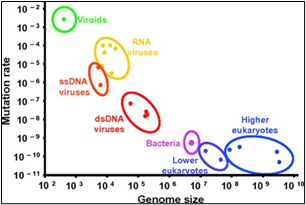
Most human coronaviruses fall under two serotypes, OC43-and 229E- surface antigen protein expressions, [1]. Fig.3, shows as per genomic length and the frequency of mutation.Viruses remain at the top with 10^-2. The new strain has a change to its spike proteins -- the regions of its outer shell that dock on our cells and infect them. The change makes it a much more efficient predator. It passes quickly from cell to cell in our bodies, copying itself at a furious pace. Baric’s experiments help to explain why the 614G strain, which first emerged in Europe in February, has quickly dominated worldwide spread,[19]. He says that the virus likely jumped out of bats and discovered a brand-new population of human hosts, with more than 7 billion of us on the planet to infect. None of us has any immune defenses against it, so we are prime targets. Viruses with genetic advantages that help them copy themselves faster and jump more quickly between hosts to survive and to cause pandemic,” says Baric, who is one of the world’s foremost experts on coronaviruses. His new study is published in the journal Science.
The new study backs up earlier research by a team of scientists led by Bette Korber, PhD, at Los Alamos National Laboratory in New Mexico. The team first noticed the rapid spread of the new strain and questioned whether the virus wasn’t evolving to become more easily passed between people. Nov. 13, 2020 -- The virus that causes COVID-19 is not the same strain of corona as it was first emerged in China, affected and killed millions of innocents globally by corona A new study shows it has changed slightly in a way that makes it more contagious to humans., Compared to the original strain, people infected with the new strain -- called 614G -- have higher viral loads in their nose and throat, though they don’t seem to get any sicker. But they are much more contagious to others.
Conclusion
Microbes and the climate change are proportionally related oscopic an outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was first reported in Wuhan City, China, in December 2019. Since then, the outbreak has grown into a global pandemic, and neither a vaccine nor a treatment for the disease, termed coronavirus disease 2019 (COVID-19), is currently available. The slow translational progress in the field of research suggests that a large number of studies are urgently required. In this context, this review explores the impact of bacteriophages on SARS-CoV-2, especially concerning phage therapy (PT). Bacteriophages are viruses that infect and kill bacterial cells. Several studies have confirmed that in addition to their antibacterial abilities, bacteriophages also show antiviral and antifungal properties. River Ganges in India, for centuries, has been revered for its “self-cleansing and special healing properties”. More than 450 million people depend on the waters of Ganges for many aspects of their life. In 1896, one of the first published works on Ganges water by Ernst Hankin, a British bacteriologist demonstrated antibacterial property of Ganges water against Vibrio cholera [1]. Further work by French microbiologist D’Herelles in the beginning of the twentieth century established that the antibacterial property of Ganges water to be due to a factor, later named “bacteriophage” [2] Virus Bacteria, (Unorganised without nucleas) Fungi, Mould and Algae (Organize with nucleas) are propagating and changing their non-pathogenic mode to pathogenic mode quickly and slowly based on their pro(unorganised) karyotic and Eu (organised, nuclear material). The somatic, reproductive and immune responsive cells of our body belong mainly by eukaryotic cells and tissues. Viral cells as prokaryon carry ss, (single stranded), or ds (double stranded) RNA or DNA and transposons. Viruses can only survive, if host for such viruses are found, [14]. For survivals,viruses adapt environment spontaneously by mutations and participate evolution. Bacteria and viruses, follow their metabolic activities, either in the form of symbiosis, partial full parasitic nature, “host–vs -graft” immune responses (IR). Bacteria at the size, of 1 micron (µ) to 10 micron (µ) irrespective to host, have their own metabolic system. On the other, viruses are fully host specific, change from non-pathogenic to pathogenic, lytic to lysogenic phases. Based on signal received from hosts, bacteriophages in case of bacteria and retro and adeno viruses in case of animal and human propagate by means of infections.As per host and environment they colonize and grow in normal and extreme, acidophilic (pH1-2), mesospheric and thermophile environments. Lambda, Psi-ex, T4 viruses share their existence by transforming DNA to RNA among adeno-, retro- and rhino, animal viruses (human, animal, bird and insect) to sustain host specific activities, [14,15]. The origin of river Ganges is known as Gomukh. Geological studies have proven that the Himalayas had emerged at a site where the Tethys Ocean once existed, as a result of collision between the Indian and Eurasian tectonic plates. Hence, Himalayas’ marine origin is known and it has been intriguing to find marine fossils high in the Himalayas [3].
References
- Nina V. Fedoroff (2012) Mc Clintock’s Challenge in the 21st century,PNAS December 11, 2012 109 (50) 20200-20203
- Starlinger P, Saedler H (1976) IS Element in Microorganism; Current Topic on Microbiology and Immunology, 75: p:111-152,
- Saedler, H, Ghosal D, and Nevers P, (1978), The role of Gene Rearrangement in Evolution, Institute for Biology III, Schaenzler Strasse, 1;7800 Freiburg, Federal Republic of Germany.
- https://onlinelibrary.wiley.com/doi/abs/10.1002/jobm.201800204
- Ruediger Schmidt (1982), Transposon; Springende Gene in Bakterien, Erbforschung Heute. Herousgegeben vom Walter Klingmueller, Verlage Chemie, Basel, Switzerland.
- Gu, H., Chen, Q., Yang, G., et al (2020). Adaptation of SARS-CoV-2 in BALB/C mice for testing vaccine efficacy. Science, 369(6511), 1603-1607.
- Shapiro JA (1979).
- Bukhari AI, Zipser D (1972).
- Georgiou, Ioannis; et.al. (2018). Retrotransposon RNA expression and evidence for retrotransposition events in human oocytes Human Molecular Genetics, Volume 18 (7).
- Brahma, N.K., Schumacher, A., Cullum, J. and Saedler H. (1982) Distribution of the Escherichia coli K-12 Insertion Sequences IS1, IS2 and IS3 among other Bacterial Species. J. Gen. Microbiol. 128: p:2229-2235.
- Brahma, N.K. (1999) Enterobacterial 026: MRHA-plasmid and its possible Genetic Transformation into Auxotrophic Escherichia coli K-12 C600 NS. Indian J. Microbial. 38 (4): p: 43-50.
- Brahma, N.K. (1999) Genetically Engineered Hybrid 5405 E. coli K-12 fimbriae in vaccination With Balb/c mice against 026: serotype EPEC diarrhea. Ind. J.Chem. Engg. 39 (1) p: 17-, Cited in British Journal catalogue (BJC)-UK
- Brahma NK (2020) Concept for Possible Development of Vaccine against Corona in Escherichia coli K-12, Yale C600 Strain, EC Clinical and Medical Case Reports 3.6 (2020): 111-116.
- Brahma NK (2020) Understanding Rapid Transmitting, Invasive, Corona Pandemics through (Aair), (Is: Tn:Is), Crispr /Cas-9 DNA Editing, Journal of Virology Research & Reports. SRC/JVRR-104.
- Greaney, A.J., Starr, T.N., Gilchuk, P., et al(2021). Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe29(1), 44-57.
- Natural Bacteriophages' Potential—A Direct Weapon Against Bacteria [16] Natallie Crowley, 2020, A Look into the Epigenetics of a Coronavirus Infection Epigenetics, March 10, Diseases & Disorders, Educationally Entertaining
- ba CP, Environmental Microbiology, (2nd Edition), Elsevier, Academic press. [Maier RM, Pepper IL, Ger Greaney, A.J., Starr, T.N., Gilchuk, P., (2009) et al. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe29(1), 44-57 (2021).
- Julia Evangelou Strait (2020)'Jumping genes' help stabilize DNA folding DNA folding patterns,Washington University School of Medicine, 24 January 2020, Phyorg.
- Chymen Chemicals (2021) News and Announcement, Concerning Variants of SARS-CoV-2, Featured Article from 2021-01-25.
- Kenneth E Bernstein and Zakir Khan, et.al (2018) Angiotensin-converting enzyme in innate and adaptive immunity PMID: 29578208 PMCID: PMC619204OI: 10.1038/nrneph.2018.15, Free PMC article
- Southern, Edwin Mellor (5 November 1975).
- Jaidip Dogra, Livodip Dogra, 2020, “Opportunistic Chlamydia in Corona, Jaipur Central Govt Health Scheme,” Pratidin Bengali newspaper, 25th May, p:1 and5.
- Braude AI, Davis, CE, Fierer j, (1981) Medical Microbiology and Infectious Diseases, Saunders Pub, International Text Book of Medicine.Page:619, D.A.J. Tyrrell Corona Virus.
- David A.J. Tyrrell and Steven H. Myint, (1980) Medical Microbiology. 4th edition,
- Jei Cui Fang Li & Zang Li Shi (2018) Origin and Evolution of Pathogenic Coronaviruses, Nature Review Microbiology, 17: p:181-192.
- Dae Gyann Ahn, et.al (2011) Interference of Ribosomal Frameshifting by antisera peptide nucleic acids suppresses SARS, Corona virus replication, Elsevier 91(1): p:1-10.
- Felix Boecker and Karin Moelling (2019) Evolution of Immune Systems from Viruses and Transposable Elements. Frontier of Microbiology, Virology, 29 Jan;
- Mitch Leslie (2020) T-Cell formed in Covid-19, patients “bode cell” for long term Immunity, Science, 14 May.
- Crispr/Cas-9 and RAG ½ T-cell diversity; Stanford Bioengineering Schools of Engineering & Medicine,
- Brahma NK (2020) Nano-Engineering, Vaccination & Diagnosis in Combating Corona: The Recent Trends”, 25.11.2020, ODTW (Oneday Technical Webinar), CHDB, by WBSC, The Institution of Engineers, India,” Feasibility Searched for Nanocoating on lung alveoli cells”. COVID-19: Bacteriophage Could Decrease Mortalityhttps://www.liebertpub.com › doi › phage.2020.0014
- Jun-2020 — The pandemic of the coronavirus disease (Covid-19) has caused the death of at least 270,000 people as of the 8th of May 2020.
- 2. Ganga Water and Phage


