Archive : Article / Volume 1, Issue 1
Case Report | DOI: https://doi.org/10.58489/2836-2187/002
Quenching of Qurom Sensing Regulated Protein of Helicobactor Pylori
1. Department of Food Science & Nutrition, Nehru Arts & Science College, Coimbatore.
2. Department of Food science and Nutrition, Mount Carmel College, Autonomous, Bengaluru, Karnataka 560052, India.
3. Sri Lakshmi Narayana Institute of Medical Sciences, Puducherry â 605 502 (affiliated to Bharath Institute of Higher Education and Research (BIHER), Chennai, Tamilnadu),
Correspondng Author: Kothandapani Sundar
Citation: Niroja R, Kumara G. K, P. Vasanthakumari, G. Jayalakshmi, K. Sundar, (2022). Quenching Of Qurom Sensing Regulated Protein of Helicobactor Pylori, Journal of Microbes and Research. 1(1). DOI: 10.58489/2836-2187/002
Copyright: © 2022 K. Sundar, this is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received Date: 2022-08-19, Received Date: 2022-08-19, Published Date: 2022-09-28
Abstract Keywords: Quorum Sensing Inhibitors (QSI), H. pylori, Hospital acquired infection, Molecular Docking, Food bioactive compounds.
Abstract
Bacteria have been shown in recent years to sense population density and change their phenotype depending on their surroundings. Quorum sensing is a phenomenon that allows bacteria to control virulence factors and biofilm formation. Because of the increasing number of therapeutic failures, Helicobacter pylori is our primary focus in this case. Obliteration of anH. pylori infection is getting decreasingly hard. Antibiotic resistance is extensively caused by biofilm conformation, a medium inH. pylori regulated by QS (signal patch- AI- 2) and regarded as a sensitive marker for quorum- seeing bacterial communication. A new result for quorum seeing attenuation was delved in this design, which involves the use of computer- supported medicine design to identify implicit quorum seeing impediments as a relief for antibiotics. Six campaigners were computationally linked as implicit QSIs from the 25 composites screened due to their high list affinity for the chemoreceptor TlpB and low toxin. This emulsion could be used to either stop or reduce bacterial communication and thus inhibit the development of QS-controlled traits by in silico method against nosocomial infection causing drug resistant H.pylori. In-vitro studies also shown that it inhibits the formation of H. pylori biofilms.
1. Introduction
Quorum sensing is an occurrence in which a When 'signal' molecules accumulate within an environment, a single cell can detect the number of bacteria present (cell density) surrounding environment, allowing the population as a whole to respond in a coordinated manner. This frequently results in signal auto induction, resulting in Signal concentrations in the surrounding environment increase rapidly Various social behaviors of bacteria have been discovered, including swarming motility, conjugal plasmid transfer, antibiotic resistance, biofilm formation, and various quorum sensing systems, common to both Gram-positive and Gram-negative bacteria., regulate many of these behaviors (Ng, Bassler 2009). The accessory gene regulator (Luxs) system has been linked to the pathogenesis of Helicobactor pylori. While luces as the control of virulence gene expression in vitro have been relatively simple, the regulation of quorum response and virulence genes in vivo has been much more difficult (Hoch 1995Other regulators influence the quorum response that relate to factors other than cell density, and it is also highly dependent on the environment in which the organism is growing. The AGR phenotype may influence biofilm-associated bacteria's behaviour and pathology. H.pylori, well contribute to the chronic nature of some biofilm-associated illnesses, according to growing evidence. Chemotaxis, or directed mobility in response to chemical cues in the environment, is a crucial characteristic of many bacterial diseases (Fuente-Nú˜nez et al., 2013). H. PH is thought to prompt H. pylori, a Gram-negative gastric pathogen, to locate itself inside the mucosa of the stomach (Schreiber et al., 2004). Dysregulated chemotaxis impairs Helicobacter pylori's ability to colonize the mouse stomach and gerbil stomach, resulting in aberrant Helicobacter pylori distribution and activation of inflammation in these tissues (Foynes et al., 2000).
Chemoreceptors, known as methyl-accepting chemotaxis proteins, mediate Chemotactic response in Escherichia coli (MCPs) (Pittman et al., 2001) is a relatively well studied phenomenon. The chemosensory ministry of Helicobacter pylori is analogous but not identical to that of Escherichia coli. Both bacteria have the core signal- transduction outfit (CheW, CheA, and CheY), but Helicobacter pylori also has three CheV proteins that contain both CheW and response- controller motifs. TlpA, TlpB, TlpC, and TlpD (firstly called HylB or HlyB) are all decoded by the Helicobacter pylori genome, though strain G27 doesn't express TlpC due to a frameshift mutation in the gene. Unlike peritrichousE. coli cells, which swim forward and tumble,H. pylori cells swim forward, reverse, and stop, as we and others have observed The endogenously produced quorum- seeing patch autoinducer- 2( AI- 2) is one of the environmental conditions thatH. pylori encounters (Rader etal., 2007). Autoinducers (AIs) are extracellular signalling motes produced by bacteria that spark quorum seeing, a type of bacterial cell – cell communication. AI attention rises as bacterial cell viscosity rises, and at critical attention, it initiates coordinated gene regulation. Several kinds of quorum- seeing systems have been linked. As AIs, gram-positive and gram-negative bacteria generally use oligopeptides and acylated homoserine lactones, independently. numerous gram-positive and gram-negative bacteria have a quorum- seeing system that generates the furanone signal AI- 2, which serves as an interspecific communication signal.
Nosocomial is an incomplete information, In spite of decades of intense debate, the transmission of Helicobacter pylori remains unclear), including how the organism leaves its host and enters the environment, where (if anywhere) the organism persists in the environment, how people become infected, and whether all people are vulnerable.infection. Circumstantial evidence suggests that direct person-to-person transmission is the most likely mode of H. pylori transmission (Schwarz et al.,2008) This can happen through fecal-oral, oral-oral, or orgastric-oral routes. The discovery of Higher rates of infection at crowded dwellings, institutions for the disabled, and households with young children suggest fecal-oral transmission. Helicobacter pylori can be isolated from the stools of adults and children that suggests fecal-oral transmission.) The ability to detect Helicobacter pylori can provide additional evidence.in the mouth suggests oral-oral transmission.
Epidemiological studies have suggested and molecular techniques have demonstrated the Nosocomial transmission of H pylori occurs through improperly cleaned and disinfected medical equipment, most commonly endoscopes. H. pylori was isolated from endoscopes and biopsy forceps that had previously been used. Following the use of pH electrodes that had not been disinfected between patients, multiple patients developed Helicobacter pylori infection as a cause of epidemic gastritis with hypochlorhydria Neelapu etal., (2014) Epidemiological studies have suggested that potentially contaminated endoscopes can transmit H. pylori. The transmission of H. pylori from patient to patient via contaminated endoscopes has been demonstrated using molecular techniques. All cases of transmission have occurred as a result of the use of ineffective disinfectants such as 70% ethanol, or 0.2 percent benzethonium chloride. H. pylori has been reported to be acquired through mouth-to-mouth transmission. According to a Endoscopy personnel had a higher prevalence of Helicobacter pylori than blood donors in the seroepidemiological study, suggesting that Helicobacter pylori is an occupational hazard based on the use of blood donors as controls. on the other hand, may have been inappropriate. Furthermore, seropositivity was not related to the number of endoscopies performed monthly or the number of years involved in endoscopy (Ryu et al., 2010)
2. Materials And Methods
2.1 Selection of bioactive compounds
To analyse docking interactions between edible bioactive compounds and quorum sensing restrained proteins of Helicobacter pylori, a molecular docking analysis was performed using AutoDock. Table 1 lists bioactive components from various foods that were chosen based on previous research findings.
Table 1: Food Source and bioactive compounds
FOOD SOURCE | BIOACTIVE COMPOUNDS |
Milk | Bovine Lactoferrin |
Green tea | (-) - epicatechin (EC) |
| (-) – epicatechin-3-gallate (ECG) |
| (-)-epigallocatechin (EGC) |
| (-)-epigallocatechin-3-gallate (EGCG) |
Zingiber officinale | 6-gingerol |
| 8-gingerol |
| 10-gingerol |
Curcuma amada | Cinnamic acid |
| Ferulic acid |
| P-coumaric acid |
| Protocatechic acid |
| Gentisic acid |
| Gallic acid |
Orange | Naringin |
| Neohesperidin |
| Hesperidin |
Grapes | Rasveratrol |
Lemon | D-Limonene |
| Limonexic acid |
Mango | Mangiferin(2-β-D-glucopyranosyl-1,3,6,7-tetrahydroxy-9H-Xanthen-9-one) |
Pumpkin, Chilli, Eggplant | Zeaxanthin |
Fish | Docohexaenoic acid |
| Eicosapentanoic acid |
Cauliflower | Quercetin |
2.2 Retrieval of 3D Structures of Target and food compounds
Protein Data Bank (PDB)
The Protein Data Bank (PDB; http://www.rcsb.org/pdb/) is a worldwide database that consist of three-dimensional structural data for biological macromolecules such as proteins and nucleic acids. The RCSB PDB website (rcsb.org) (Rose et al.,2015). The RCSB Protein Data Bank: Views of structural biology for basic and applied research and education. Nucleic Acids Res includes a variety of Tools for structure querying, browsing, analysis, and molecular visualisation are available. Users can use either the top menu bar search, which makes use of PDB ID, name, sequence, and ligand SMILES, or they can use Advanced Search to create complex search combinations. PDB databases are arranged into hierarchical trees using external classification and annotation systems for browsing and searching. (e.g., Membrane Proteins, Gene Ontology, Enzyme Classification) Exploration of 3D structures, structure/sequence correlation, and correspondences between the two is possible with visualisation options. The Protein Feature View, for example, provides a graphical comparison of a PDB sequence with UniProt and other annotations displayed in different tracks.
These structures are available and have been submitted by scientists using biological approaches like as X-ray crystallography, NMR spectroscopy, and cryo-electron microscopy. The high-resolution three-dimensional structures of the target protein (PDB ID: 1J6X) used in this paper are from the PDB database and we also used the structure of known anti-quorum sensing compound bounded to our preferred targets.
PubChem
Millions of compound structures and illustrative datasets can be freely downloaded through FTP from PubChem (https://pubchem.ncbi.nlm.nih.gov), a public chemical information database at the National Center for Biotechnology Information (NCBI) (Kim et al., 2016). Substance, Compound, and Bioassay are three interconnected databases created by PubChem. We're using the PubChem database to get the structures of 25 food-derived compounds. The downloaded structures are in SDF format, which can be translated to MOL2 format for docking. PubChem divides this massive Data collected in three interconnected databases: Substance, Compound, and BioAssay: Compound Database stored in the Substance Database (https://www.ncbi.nlm.nih.gov/pcsubstance) stores information provided by depositors. Compound Database retrieves chemical structures, which are then stored in the Substance database. (Wang etal.,2017) (https://www.ncbi.nlm.nih.gov/pccompound). Descriptions of biological assays on chemical substances are stored in the BioAssay database (https://www.ncbi.nlm.nih.gov/pcassay).
2.3. Molecular Docking of Bioactive Compounds to Target by AutoDock
The interaction between molecules underpins the majority of biological processes. These molecules might be proteins, enzymes, or medicines, and the optimal result is which is the consequence of a convenient interaction between them. Docking is the software's prediction of these interactions. Docking is a molecular modelling method that predicts how well a protein interacts with a ligand molecule in a basic description. Docking procedures are quite beneficial in a variety of biological research domains, and we definitely want some docking software to carry out these activities. The Auto Dock web site is devoted to small molecule docking on target proteins. It offers simple docking methods that allow the target protein and ligand structures to be automatically constructed for docking.
Furthermore, the docking runs do not require any computational resources from the user; the server handles all of the calculations. Docking experiments were carried out with the help of the programme AUTODOCK 4. This programme starts with a ligand molecule in any conformation, orientation, or position and uses both simulating annealing and genetic algorithms to find favourable dockings in a proteinbinding site. The protein and ligands were prepared using the AutoDockTools (ADT) programme, which is availableWe adapted the Python Molecular Viewer extension suite to dock macromolecules, and used a three-dimensional crystal structure of HpFabZ (PDB entry: 3CF9), resolved at 2.6, to perform docking studies. We kept only the A, B, and C chains of the hexamer, whereas we combined and deleted the D, E, and F chains. This macromolecule has two active sitesAside from this, both binding and interaction sites lie within the C side chain and the tunnel between the A and B chains. The crystal's existing molecules of ligands were all removed to facilitate docking, along with the crystallographic water molecules. Polar hydrogens were then added, followed by Kollman Units, Atomic solvation parameters and charges were then assigned, Finally, atomic solvation parameters and charges were calculated. To perform molecular docking, the GA-LS method was used. The GA parameters were Maximum 250,000 energy evaluations; maximum 27,000 generations; and mutation and crossover rates of 0.02 and 0.8, respectively. For local search, sub-Solis & Wets parameters were used, and 300 iterations of a Solis & Wets local search were imposed. were limited to 50. Cygwin was used to run the Autogrid and Autodock computations Structures generated after docking were grouped based on a tolerance of 1 A ° RMSD from the lowest-energy structure. ADT was used to investigate hydrogen bonding and hydrophobic interactions between docked potent agents and macromolecules (Version 1.50).
2.4. In-vitro studies for quorum sensing inhibition (QSI) activity
2.4.1. Minimum inhibitory concentration (MIC) of Neohesperidin and Naringin against H.pylori
The MIC of Neohesperidin and naringin against H.pylori was determined using the Clinical and Laboratory Standards Institute, USA (2006). Overnight, a bacterial culture was inoculated into 20 ml of LB medium with various concentrations of Neohesperidin and naringin ranging from 0.1 mg to 100 mg and incubated at 37°C for 24 hours. The absorbance of the medium was measured at 600nm before and after incubation. The MIC was the lowest concentration of Neohesperidin and naringin that inhibited growth.
2.4.2 Reduction in exoplysaccharide production
Cells adhered to the test tube walls were harvested at an early stationary phase by centrifugation at 8000 rpm for 30 minutes at 4°C to obtain crude EPS. The filtrate was passed through a 0.22 mm membrane and incubated overnight at 2°C with three volumes of cold ethanol to precipitate the EPS. Precipitated EPS was collected and dissolved in 1 ml of deionized water after centrifugation at 8000 rpm for 30 minutes. The EPS was measured using the phenol-sulfuric acid method (Huston et al., 2004). The effects of Neohesperidin and Naringin on EPS reduction were investigated at sub-MIC levels.
2.4.3 Inhibition of H.pylori biofilms by microtitre plate assay
The biofilm inhibition assay was carried out using a microtitre plate assay. In microtitre plates, 1% of an overnight H.pylori culture was inoculated with and without Neohesperidin and Naringin and incubated at 37°C. After incubation, planktonic H.pylori cells were carefully rinsed with sterile distilled water, and surface adhered biofilms were stained with 0.2 percent crystal violet for 10 minutes. Excess stain was drained, and crystal violet in biofilms was dissolved in 95 percent ethanol. The biomass of H.pylori biofilms was quantified using a microtitre plate reader to measure the crystal violet intensity at OD650nm (Limsuwan and Voravuthikunchai, 2008).
3. Result
As as result twenty-four bioactive compounds from different food-based molecules are collected from PubChem database and are subjected to molecular docking studies using AutoDock. The crystallographic structures of LuxS protein with PDB ID: 1J6X is used as target.The molecular docking data were obtained from different edible bioactive compounds and its interactions between the secondary metabolites and the target receptors such as LuxS protein. These results of in silico molecular dockings studies evidenced the possible regulatory role of the phytochemicals present in the natural resources. The compound that is extensively used for inhibiting the biofilm formation of H. pylori was docked for the comparison purpose. The results obtained from AutoDock had been analyzed full fitness and ΔG Kcal/mol is noted in the table 2.
Table 2: Flexible Docking of LuxS (PDB: 1J6X) with Top Ranked Bioactive Compounds
| Compound ID | Compound Name | Ligand Efficiency [kcal/mol] | Dock Score [kcal/mol] | Binding Energy [kcal/mol] |
| 30231 | Neohesperidin | -0.260 | -11.192 | -56.795 |
| 9894584 | Naringin | -0.236 | -9.665 | -56.541 |
| 10621 | Hesperidin | -0.195 | -8.377 | -53.864 |
| 102025303 | Epigallocatechin3gallate | -0.142 | -7.927 | -65.596 |
| 12795889 | Epicatechin3gallate | -0.137 | -7.254 | -54.290 |
| 5281647 | Mangiferrin | -0.216 | -6.488 | -42.499 |
| 3473 | 6gingerol | -0.299 | -6.288 | -38.792 |
| 65064 | Epigallocatechin | -0.190 | -6.284 | -52.600 |
| 168114 | 8gingerol | -0.265 | -6.088 | -40.096 |
| 107905 | Epicatechin | -0.188 | -6.022 | -49.086 |
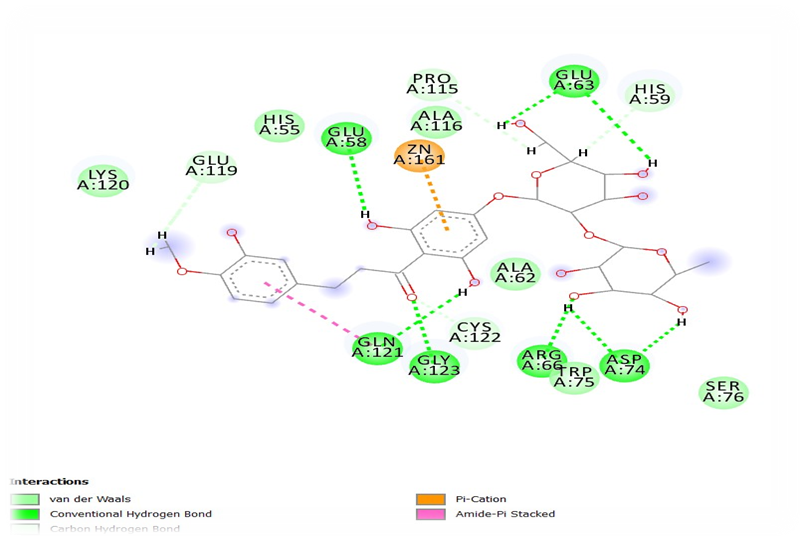
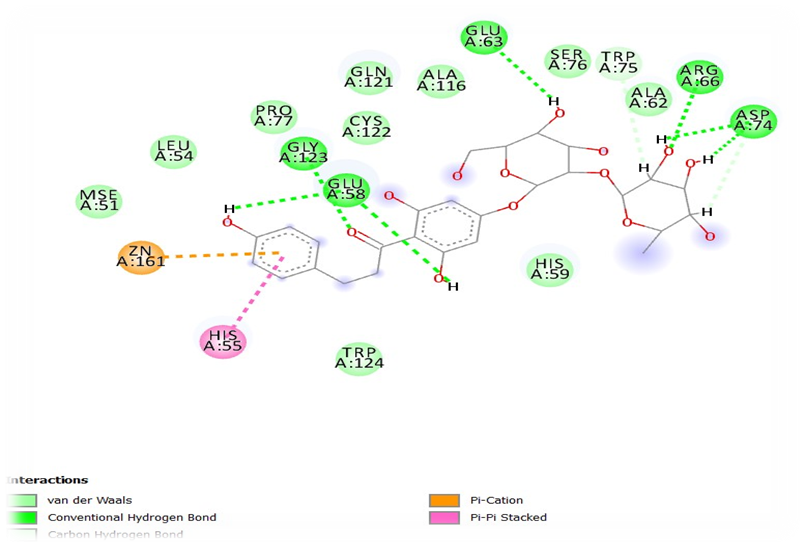
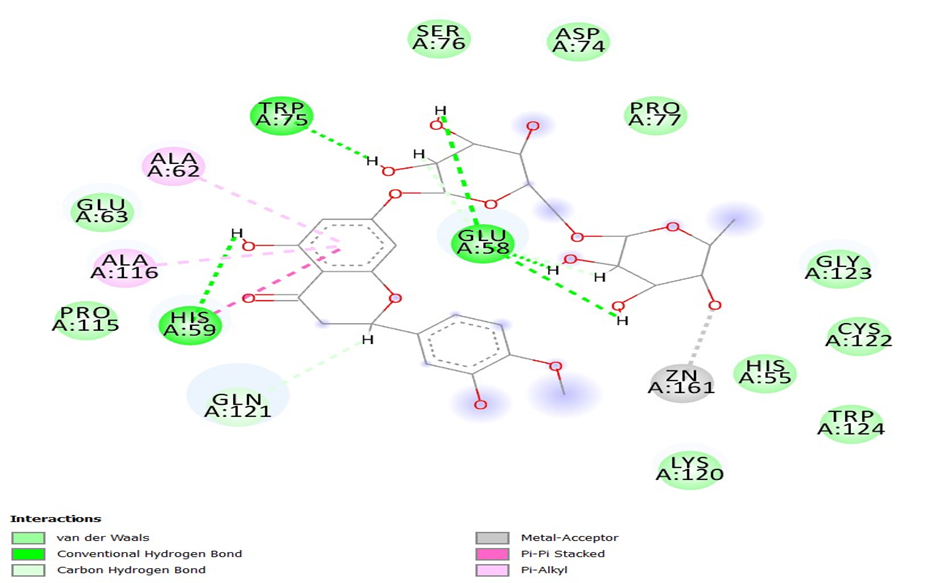
The best two compounds Neohesperidin and naringin were carried forwards for the in-vitro studies based on the in-silico results. Minimum inhibitory concentration (MIC) of EGCG and naringin was studied against H.pylori and showed 125µg/ml and 50 mg/ml respectively.
Inhibition of EPS production and biofilm formation by Neohesperidin at sub-MIC levels were represented in figure 4. Concentration dependent reductions in EPS production and H.pylori biofilm formation of naringin were shown in figure 5. Neohesperidin inhibited maximum upto 48% EPS production in H.pylori and naringin showed 41
4. Discussion
The discovery of Helicobacter Pylori 20 years ago changed the diagnosis and treatment of gastroduodenal disease. In this review, we examine the epidemiology and transmission of Helicobacter pylori as well as its role in upper gastrointestinal tract disease. We also discuss diagnostic approaches, indications for therapy, and therapeutic efficacy measures.
The non-treating of Helicobacter pylori infection can lead to acute transmissible diseases, which range from sores and ulcers to potentially fatal cancers. Previously, Helicobacter pylori infections were treated with a combination of two or more antibiotics. MDR in Helicobacter pylori has been linked to the upregulation of efflux pump genes, including hp1145, AI-2 functions as a quorum sensing component in Helicobacter pylori infections, and a decline in the effectiveness of antibiotics in Helicobacter pylori infections has been linked to antibiotic resistance.
The previous study demonstrated has been shown that EGCG has cytoprotective actions against Helicobacter pylori-associated cytotoxicity, which are distinguished by enhancement of cell proliferation, DNA damage attenuation, and anti-inflammatory effects. There is evidence, albeit limited, that some blue-green flavonoids, like ginsenoside, tannins, or saponins, behave in a biphasic manner based on their biological effects/cytoprotection or cytotoxicity, antioxidant or pro-oxidant, or DNA damaging or DNA protecting. (Hu et al.,2001). From my studies the molecular docking has been done with edible bioactive compound that interact with protein molecules as a result it shows the best docking score with 3 compounds as neohesperidin, naringin, hesperidin the binding energy with those molecules are high compared to another bioactive compound. The compound which has antimicrobial activities will be taken for the in-vitro studies.
5. Conclusion
Final result of the current,in silico study for determining the anti-quorum sensing activity of Twenty-four different bioactive compounds that are commonly found in various food sources, against H.pylori receptor protein LuxS, found that 10 molecules had similar high docking scores with the known inhibitor and thus can be used as potential inhibitors of H.pylori biofilm formation. The compounds that showed the high fitness scores have to be suggested for further in vitro and in vivo studies to ensure its potential. Further gene expression and molecular interaction studies are also needed for understanding more about these quorum quenching molecules and their mode of action in depth. The potential quorum sensing inhibitors can be developed as new effective drug candidates and may help in effective treatment along with the current antibiotics.
6. Significance of The Study
Helicobacter pylori is recognized as the major cause of gastric disease around the world and is rising in prevalence. According to conventional theory, Helicobacter pylori can be transmitted in three ways: iatrogenic, oral-oral, and faecal-oral (gomes et al.,2004). It’s mainly caused to the childrens and it may lead to the death when its left untreated.
Author Contributions
All the authors contributed equally for this research work.
Funding source
Nil
Conflict of interest
The authors have no conflict of interest.
References
- B.C. Gomes; E.C.P. De Martinis (2004). The significance of Helicobacter pylori in water, food and environmental samples. , 15(5), 397–403. doi:10.1016/s0956-7135(03)00106-3
- de la Fuente-Nú˜nez C, Reffuveille F, Fernandez L, REW H. Bacterial biofilm development as a multicellular adaptation: antibiotic resistance and new therapeutic strategies. Curr Opi Microbiol. 2013. DOI: 10.1016/j.mib.2013.06.013
- Dong YH, Xu JL, Li XZ, Zhang LH.2000. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci.USA 97:3526–31. DOI: 10.1073/pnas.97.7.3526
- Foynes S., Dorrell N., Ward S. J., Stabler R. A., McColm A. A., Rycroft A. N., Wren B. W. (2000). Helicobacter pylori possesses two CheY response regulators and a histidine kinase sensor, CheA, which are essential for chemotaxis and colonization of the gastric mucosa. Infect Immun 68, 2016–2023. 10.1128/IAI.68.4.2016-2023.2000. DOI: 10.1128/IAI.68.4.2016-2023.2000
- Furukawa A, Oikawa S, Murata M, Hiraku Y, Kawanishi S. (-)-Epigallocatechin gallate causes oxidative damage to isolated and cellular DNA. Biochem Pharmacol 2003;66:1769–78. DOI: 10.1016/s0006-2952(03)00541-0
- Harder, E.; Damm, W.; Maple, J.; Wu, C.; Reboul, M.; Xiang, J. Y.; Wang, L.; Lupyan, D.; Dahlgren, M. K.; Knight, J. L.; Kaus, J. W.; Cerutti, D. S.; Krilov, G.; Jorgensen, W. L.; Abel, R.; Friesner, R. A. OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J. Chem. Theory Comput. 2016, 12, 281−296. doi: 10.1016/j.jmb.2016.11.021
- Hu C, Kitts DD. Evaluation of antioxidant activity of epigallocatechin gallate in biphasic model systems in vitro. Mol Cell Biochem 2001;218:147– 55. https://doi.org/10.1023/A:1007220928446
- Neelapu, N. R. R., Nammi, D., Pasupuleti, A. C. M., & Surekha, C. (2014). Helicobacter pylori induced gastric inflammation, ulcer, and cancer: A pathogenesis perspective. Interdisciplinary Journal of Microinflammation, 1, 113. doi: 10.4172/2381-8727.1000113
- Neelapu, N. R. R., Nammi, D., Pasupuleti, A. C. M., & Surekha, C. (2014). doi: 10.4172/2381-8727.1000113
- Ng WL, Bassler BL 2009. Bacterial quorum-sensing network architectures. doi: 10.1146/annurev-genet-102108-134304
- Pittman M. S., Goodwin M., Kelly D. J. (2001). Chemotaxis in the human gastric pathogen Helicobacter pylori: different roles for CheW and the three CheV paralogues, and evidence for CheV2 phosphorylation. DOI: 10.1099/00221287-147-9-2493
- Rose P.W., Prlić A., Bi C., Bluhm W.F., Christie C.H., Dutta S., Green R.K., Goodsell D.S., Westbrook J.D., Woo J.et al. . The RCSB Protein Data Bank: Views of structural biology for basic and applied research and education. Nucleic Acids Res. 2015; 43:D345–D356. doi: 10.1093/nar/gku1214.
- Ryu KH, Yi SY, Na YJ, et al. Reinfection rate and endoscopic changes after successful eradication of H. pylori. World. J. Gastroenterol. 2010. DOI: 10.3748/wjg.v16.i2.251
- Sastry, G. M.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput.-Aided Mol. Des. 2013, 27, 221−234. doi: 10.1007/s10822-013-9644-8
- Schreiber S., Konradt M., Groll C., Scheid P., Hanauer G., Werling H. O., Josenhans C., Suerbaum S. (2004). The spatial orientation of Helicobacter pylori in the gastric mucus. Proc Natl Acad Sci U S A 101, 5024–5029. doi: 10.1073/pnas.0308386101
- Schwarz S, Morelli G, Kusecek B, Manica A, Balloux F, Owen R.J, Graham D.Y, van der M.S, Achtman M, Suerbaum S. Horizontal versus familial transmission of H. pylori. PLoS Pathogens. 2008 doi: 10.23750/abm.v89i8-S.7947
- van der Woude H, Gliszczynska-Swiglo A, Struijs K, Smeets A, Alink GM, Rietjens IM. Biphasic modulation of cell proliferation by quercetin at concentrations physiologically relevant in humans. Cancer Lett 2003;200:41–7 DOI: 10.1016/s0304-3835(03)00412-9
- Wang Y., Bryant S.H., Cheng T., Wang J., Gindulyte A., Shoemaker B.A., Thiessen P.A., He S., Zhang J. PubChem BioAssay: 2017 update. Nucleic Acids Res. 2017; 45:D955–D963. DOI: 10.1093/nar/gkw1118
- Waters CM, Bassler BL. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol 21:319 –346. https://doi.org/10.1146/annurev.cellbio.21.012704.131001.
- Williams P, Camara M 2009. Quorum sensing and environmental adaptation: A tale of regulatory networks and multifunctional signal molecules


